Researchers from the RIKEN Global Research Cluster in Japan have developed a potential mouse model for the genetic disorder known as an NGLY1 deficiency. Researchers showed that while a complete knockout of the Ngly1 gene in mice leads to death just before birth, a second knockout of another gene called Engase can allow mice to survive. The doubled-deletion mice also had symptoms similar to those of people with NGLY1-deficiency. This indicates that these mice could be useful for testing potential therapies.
NGLY1-deficiency is a rare and relatively newly discovered genetic disorder. The symptoms are severe, and include delayed development, disordered movement, low muscle tone and strength, and the inability to produce tears. Understanding how lack of NGLY1 leads to these symptoms is critical when considering targets for therapeutic interventions. Creating useful animal models of the disease is therefore equally important.
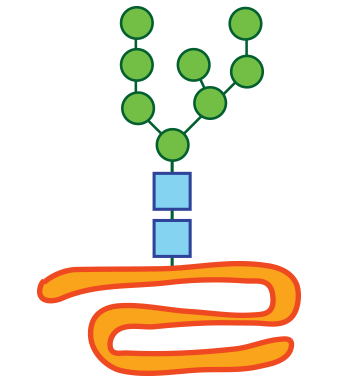
Diagram of a misfolded glycoprotein.
The RIKEN team has already had some success studying the consequences of Ngly1 deficiency in cultured animal cells. The Ngly1 gene codes for an enzyme involved in the deglycosylation of proteins that are scheduled to be recycled because they did not fold properly. Their previous research showed that when Ngly1 was absent, sugars normally removed by Ngly1 were improperly removed by another enzyme called ENGase. This led to the accumulation of protein that could not be degraded. Knocking out the ENGase gene led to normal protein degradation.
In the current study, the researchers brought their research out from the dish and into animals. First they examined the effects of knocking out Ngly1 in mice. They found that when mice lacked both Ngly1 genes—one from each parent—they always died just before birth. However, a double knockout of both Ngly1 and ENGase resulted in mice that survived after birth, but not for very long.
This positive result was actually unexpected. “We thought that ENGase acted further downstream to Ngly1 in the catabolism of glycoproteins” notes team leader Tadashi Suzuki, “and were surprised when the double knockout was able to suppress the lethality of Ngly1-KO mice. If ENGase was merely an enzyme downstream of Ngly1, nothing should have happened. This was truly a case of serendipity.”
Although the double knockout mice survived, they shared several defects that are similar to symptoms observed in people with NGLY1-deficiency. As the mice aged, they developed characteristics such as bent spines, trembling, limb-clasping and shaking, and by 45 weeks, the survival rate was reduced to 60%. These mice could therefore be useful model mice for developing treatments for the human genetic disorder.
Another factor affecting survival and symptoms turned out the be the Genetic background of the mice. When mice were crossed with an outbred strain, the single Ngly1 knockout proved less lethal, and the double knockout proved to be even more helpful. These mice only showing hind-limb clasping after 30 weeks.
These findings show that the biological processes involved are quite complicated. Even so, preventing ENGase from acting clearly alleviates symptoms of Ngly1 deficiency in mice. Figuring out which aspects of the genetic background help reduce symptoms is perhaps a long-term goal.
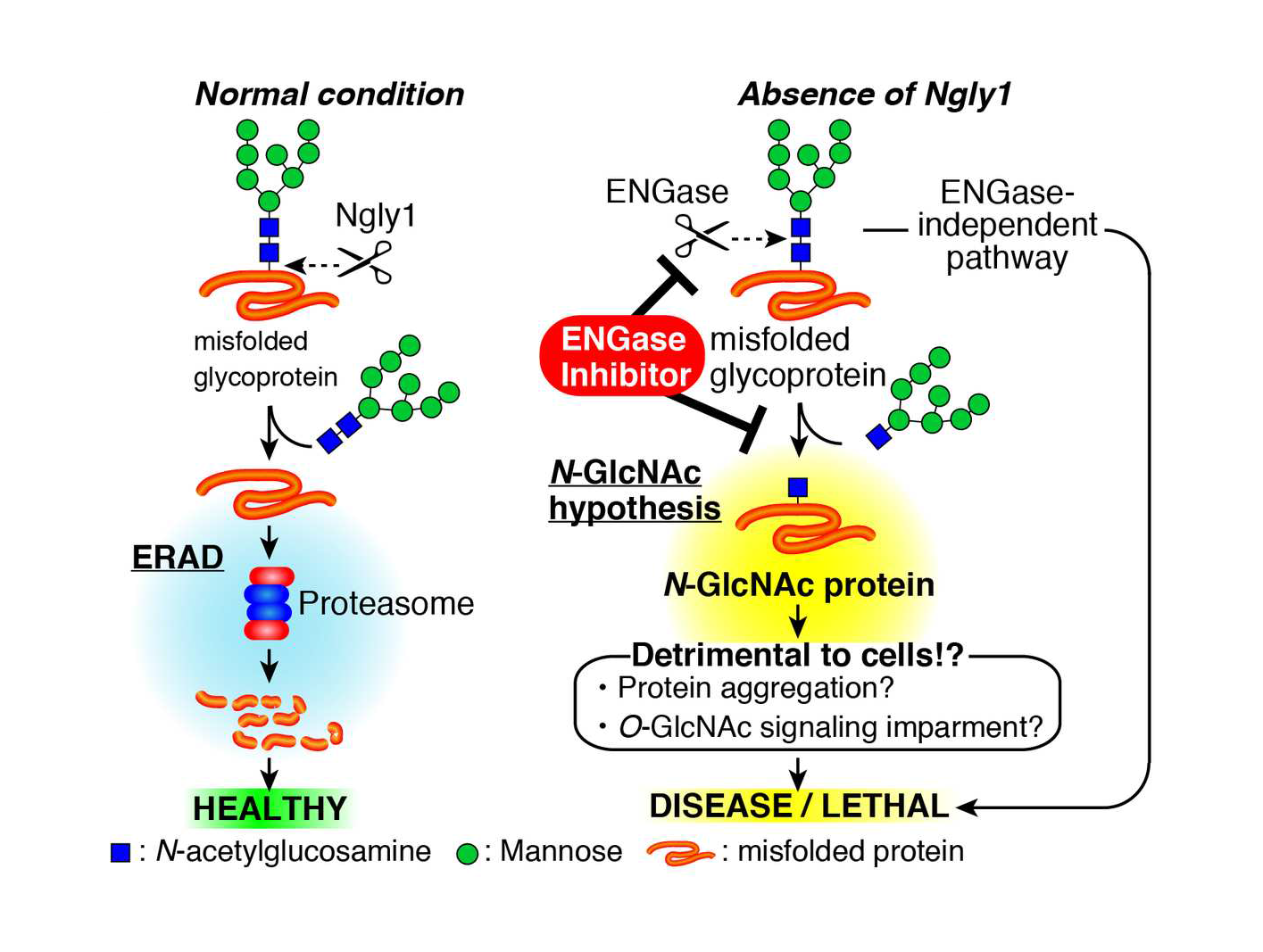
Schematic representation of an N-GlcNAc hypothesis and therapeutic treatment of NGLY1-deficiency based on ENGase inhibition. In the absence of Ngly1, ENGase acts on some portions of unfolded glycoproteins to form N-GlcNAc proteins. The presence of an excess of N-GlcNAc proteins somehow results in detrimental effects on cells/mice.
Although the condition was only recently discovered, research into NGLY1-deficiency has been facilitated through efforts from the Grace Science Foundation, and the RIKEN team has recently begun a collaboration with Takeda Pharmaceuticals and T-CiRA at Kyoto University.
“For now, the next step,” says Suzuki, “will be to isolate an in vivo inhibitor for ENGase and determine whether it can improve the symptoms related to NGLY1-deficiency. As we do not know much about the pathophysiology of the disorder, this might help us find potential targets for therapy, but also might lead to a better understanding of other diseases. Such chains of unexpected results are the beauty of basic science!”
More information about NGLY1-deficiency can be found here.
Researchers from the RIKEN Global Research Cluster in Japan have developed a potential mouse model for the genetic disorder known as an NGLY1 deficiency. Researchers showed that while a complete knockout of the Ngly1 gene in mice leads to death just before birth, a second knockout of another gene called Engase can allow mice to survive. The doubled-deletion mice also had symptoms similar to those of people with NGLY1-deficiency. This indicates that these mice could be useful for testing potential therapies.
NGLY1-deficiency is a rare and relatively newly discovered genetic disorder. The symptoms are severe, and include delayed development, disordered movement, low muscle tone and strength, and the inability to produce tears. Understanding how lack of NGLY1 leads to these symptoms is critical when considering targets for therapeutic interventions. Creating useful animal models of the disease is therefore equally important.

Diagram of a misfolded glycoprotein.
The RIKEN team has already had some success studying the consequences of Ngly1 deficiency in cultured animal cells. The Ngly1 gene codes for an enzyme involved in [itg-tooltip tooltip-content=”<p><strong>Glycosylation: </strong>the attachment of a sugar chain (glycad) to a protien or lipid.</p><br/><p><strong>Deglycoslaiton:</strong> The removal of the glycad.</p>”]deglycosylation[/itg-tooltip] of proteins that are scheduled to be recycled because they did not fold properly. Their previous research showed that when Ngly1 was absent, sugars normally removed by Ngly1 were improperly removed by another enzyme called ENGase. This led to the accumulation of protein that could not be degraded. Knocking out the ENGase gene led to normal protein degradation.
In the current study, the researchers brought their research out from the dish and into animals. First they examined the effects of knocking out Ngly1 in mice. They found that when mice lacked both Ngly1 genes—one from each parent—they always died just before birth. However, a double knockout of both Ngly1 and ENGase resulted in mice that survived after birth, but not for very long.
This positive result was actually unexpected. “We thought that ENGase acted further downstream to Ngly1 in the catabolism of glycoproteins” notes team leader Tadashi Suzuki, “and were surprised when the double knockout was able to suppress the lethality of Ngly1-KO mice. If ENGase was merely an enzyme downstream of Ngly1, nothing should have happened. This was truly a case of serendipity.”
Although the double knockout mice survived, they shared several defects that are similar to symptoms observed in people with NGLY1-deficiency. As the mice aged, they developed characteristics such as bent spines, trembling, limb-clasping and shaking, and by 45 weeks, the survival rate was reduced to 60%. These mice could therefore be useful model mice for developing treatments for the human genetic disorder.
Another factor affecting survival and symptoms turned out the be the [itg-tooltip tooltip-content=”<p>Genetic background: <span style=&aquot;color: #666666; font-family: ‘Open Sans’, Arial, sans-serif;&aquot;>the genotypes of all genes related to the target genese (in this case </span><em style=&aquot;color: #666666; font-family: ‘Open Sans’, Arial, sans-serif;&aquot;>Ngly1</em><span style=&aquot;color: #666666; font-family: ‘Open Sans’, Arial, sans-serif;&aquot;> and </span><em style=&aquot;color: #666666; font-family: ‘Open Sans’, Arial, sans-serif;&aquot;>Engase)</em><span style=&aquot;color: #666666; font-family: ‘Open Sans’, Arial, sans-serif;&aquot;>, which likely affect how they function.</span></p>”]genetic background[/itg-tooltip] of the mice. When mice were crossed with an outbred strain, the single Ngly1 knockout proved less lethal, and the double knockout proved to be even more helpful. These mice only showing hind-limb clasping after 30 weeks.
These findings show that the biological processes involved are quite complicated. Even so, preventing ENGase from acting clearly alleviates symptoms of Ngly1 deficiency in mice. Figuring out which aspects of the genetic background help reduce symptoms is perhaps a long-term goal.

Schematic representation of an N-GlcNAc hypothesis and therapeutic treatment of NGLY1-deficiency based on ENGase inhibition. In the absence of Ngly1, ENGase acts on some portions of unfolded glycoproteins to form N-GlcNAc proteins. The presence of an excess of N-GlcNAc proteins somehow results in detrimental effects on cells/mice.
Although the condition was only recently discovered, research into NGLY1-deficiency has been facilitated through efforts from the Grace Science Foundation, and the RIKEN team has recently begun a collaboration with Takeda Pharmaceuticals and T-CiRA at Kyoto University.
“For now, the next step,” says Suzuki, “will be to isolate an in vivo inhibitor for ENGase and determine whether it can improve the symptoms related to NGLY1-deficiency. As we do not know much about the pathophysiology of the disorder, this might help us find potential targets for therapy, but also might lead to a better understanding of other diseases. Such chains of unexpected results are the beauty of basic science!”
More information about NGLY1-deficiency can be found here. ?
[vifblike]
Further reading:
[yuzo_related]