A research team at the RIKEN Center for Biosystems Dynamics Research (BDR) in Japan has succeeded in experimentally evolving the common bacteria Escherichia coli under pressure from a large number of individual antibiotics. In doing so, they were able to identify the mechanisms and constraints underlying evolved drug resistance. Their findings, published in the scientific journal Nature Communications, can be used to help develop drug-treatment strategies that minimize the chance that bacteria will develop resistance.
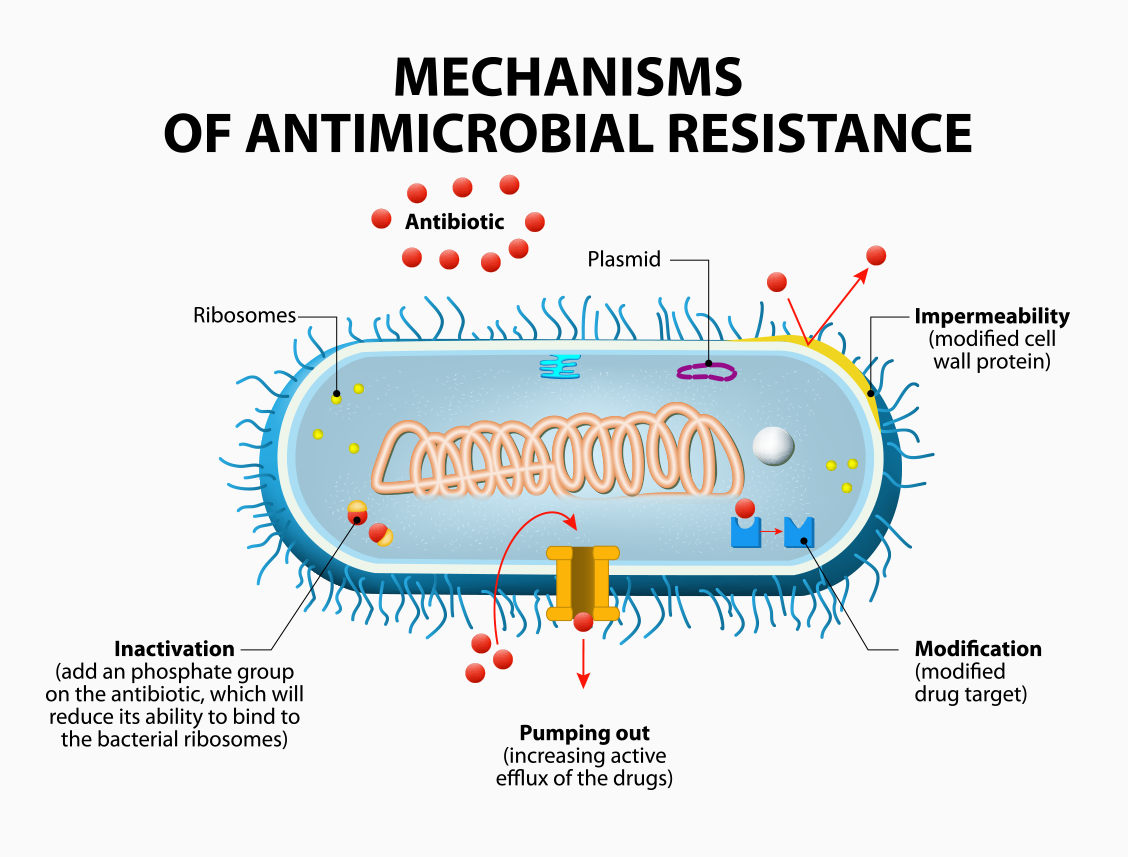
Counteracting multidrug-resistant bacteria is becoming a critical global challenge. It seems that every time we develop new antibiotics, novel antibiotic-resistant bacteria emerge during clinical use. To win this cat-and-mouse game, we must understand how drug resistance evolves in bacteria. Naturally, this process is very complicated, involving numerous changes in genome sequences and cellular states. Therefore, a comprehensive study of resistance dynamics for large numbers of antibiotics has never been reported.
 “Laboratory evolution combined with genomic analyses is a promising approach for understanding antibiotic resistance dynamics,” explains Tomoya Maeda, a researcher at RIKEN BDR who led this study. “However, laboratory evolution is highly labor-intensive, requiring serial transfer of cultures over a long period and a large number of parallel experiments.” Additionally, Maeda says that identifying the genes that allow resistance to antibiotics is not always easy because of the large number of genetic features that are contained within the data.
“Laboratory evolution combined with genomic analyses is a promising approach for understanding antibiotic resistance dynamics,” explains Tomoya Maeda, a researcher at RIKEN BDR who led this study. “However, laboratory evolution is highly labor-intensive, requiring serial transfer of cultures over a long period and a large number of parallel experiments.” Additionally, Maeda says that identifying the genes that allow resistance to antibiotics is not always easy because of the large number of genetic features that are contained within the data.
To overcome these limitations, the team developed an automated robotic culture system that allowed them to successfully perform high-throughput laboratory evolution of E. coli for more than 250 generations under pressure from 95 different antibiotics. With this new ability, they were able to quantify changes in the bacteria’s transcriptome—the set of all messenger RNAs and their transcripts, which is the record of which genes are actually expressed. As a result, the system produced resistance profiles for 192 of the evolved strains. The researchers also developed a machine-learning method for analyzing this large amount of data, allowing them to identify both novel and well-known genes that contribute to the prediction of resistance evolution.
“We found that E. coli’s evolutionary dynamics is attributable to a relatively small number of intracellular states, indicating that it is likely equipped with only a limited number of strategies for antibiotic resistance,” says Maeda. By being able to quantify the constraints that affect evolution of antibiotic resistance in E. coli, the team hopes they can predict, and thus control, antibiotic resistance.
For example, by using this new system, they were able to test 2162 pairs of drug combinations and discovered 157 pairs that have the potential to suppress antibiotic resistance acquisition in E. coli. As Maeda says, “We believe that our results can be applied to the development of alternative strategies for suppressing the emergence of drug-resistant bacteria.” ✅
Counteracting multidrug-resistant bacteria is becoming a critical global challenge. It seems that every time we develop new antibiotics, novel antibiotic-resistant bacteria emerge during clinical use. To win this cat-and-mouse game, we must understand how drug resistance evolves in bacteria. Naturally, this process is very complicated, involving numerous changes in genome sequences and cellular states. Therefore, a comprehensive study of resistance dynamics for large numbers of antibiotics has never been reported.
 “Laboratory evolution combined with genomic analyses is a promising approach for understanding antibiotic resistance dynamics,” explains Tomoya Maeda, a researcher at RIKEN BDR who led this study. “However, laboratory evolution is highly labor-intensive, requiring serial transfer of cultures over a long period and a large number of parallel experiments.” Additionally, Maeda says that identifying the genes that allow resistance to antibiotics is not always easy because of the large number of genetic features that are contained within the data.
“Laboratory evolution combined with genomic analyses is a promising approach for understanding antibiotic resistance dynamics,” explains Tomoya Maeda, a researcher at RIKEN BDR who led this study. “However, laboratory evolution is highly labor-intensive, requiring serial transfer of cultures over a long period and a large number of parallel experiments.” Additionally, Maeda says that identifying the genes that allow resistance to antibiotics is not always easy because of the large number of genetic features that are contained within the data.
To overcome these limitations, the team developed an automated robotic culture system that allowed them to successfully perform high-throughput laboratory evolution of E. coli for more than 250 generations under pressure from 95 different antibiotics. With this new ability, they were able to quantify changes in the bacteria’s transcriptome—the set of all messenger RNAs and their transcripts, which is the record of which genes are actually expressed. As a result, the system produced resistance profiles for 192 of the evolved strains. The researchers also developed a machine-learning method for analyzing this large amount of data, allowing them to identify both novel and well-known genes that contribute to the prediction of resistance evolution.
“We found that E. coli’s evolutionary dynamics is attributable to a relatively small number of intracellular states, indicating that it is likely equipped with only a limited number of strategies for antibiotic resistance,” says Maeda. By being able to quantify the constraints that affect evolution of antibiotic resistance in E. coli, the team hopes they can predict, and thus control, antibiotic resistance.
For example, by using this new system, they were able to test 2162 pairs of drug combinations and discovered 157 pairs that have the potential to suppress antibiotic resistance acquisition in E. coli. As Maeda says, “We believe that our results can be applied to the development of alternative strategies for suppressing the emergence of drug-resistant bacteria.” ✅
Further reading
Maeda et al. (2020) High-throughput laboratory evolution reveals evolutionary constraints in Escherichia coli. Nat Comm. doi: 10.1038/s41467-020-19713-w
Further reading
Maeda et al. (2020) High-throughput laboratory evolution reveals evolutionary constraints in Escherichia coli. Nat Comm. doi: 10.1038/s41467-020-19713-w