The paper reported the development of a new type of microscopy by a group led by Eric Hetzig at the Howard Hughes Medical Institute, which allows extremely rapid, precise, and relatively non-destructive 3D imaging of living cells. The technology essentially uses a laser to illuminate a thin slice of a sample, and that slice is then imaged by a device set perpendicular to the plane of the slice. By taking slice after slice, a 3D image of the sample can be constructed.
We recently caught up with Yuko and asked her some questions about the prize.
First of all, congratulations! We’d like to start by asking, what was your role in the collaboration?
The group was led by Eric Hetzig, who won a Nobel Prize in 2014 for his work on super-resolved fluorescence microscopy. In the paper last year we demonstrated a new imaging method, called lattice light-sheep microscopy. What I did was provide the group with [itg-tooltip tooltip-content=”<p><strong>A HeLa cell </strong> /ˈhiːlɑː/, also Hela or hela cell, is a cell type in an immortal cell line used in scientific research. (Source: Wikipedia)</p>”]HeLa cells[/itg-tooltip] expressing EB1-[itg-tooltip tooltip-content=”<p><strong>GFP</strong> is the abbreviation for green florescent protein. In research, it is often genetically inserted into cells and attached to the production of other cellular companant as a way to visialize elements within a cell.</p>”]GFP[/itg-tooltip], a marker for microtubule growth, and H2B-[itg-tooltip tooltip-content=”<p><strong>RFP</strong> is the abbreviation for red florescent protein, which is used similarly to GFP.</p>”]RFP[/itg-tooltip] visualizing chromosomes. I asked them to use the method to take 3D time-lapse movies. Initially, they tried using a prototype called a “Bessel beam microscope,” but it was not successful so they continued to improve the technology and finally developed the lattice light-sheet microscope and succeeded in taking marvelous 3D movies. During the data collection and analysis, we were continuously discussing how we could succeed.
How did you become involved with Eric Hetzig’s group, and did you feel it was valuable to be in a collaboration?
It’s a long story. For more than ten years, I had been looking for novel technologies that would make it possible to create 3D images of living cells at high spatiotemporal resolution to visualize the dynamics of molecules.
In the mid-2000s, two novel technologies became available: light sheet microscopy and super-resolution microscopy. But neither of these techniques could satisfy my needs, because conventional light sheet microscopes were optimized for large specimens and the spatial resolution was too low, while the frame speed of super-resolution microscopy was too slow. So I tried to develop a fast-scanning high-resolution confocal microscope, but the acquisition speed was not yet satisfactory for examining subcellular events at sub-second rate.
In 2011, to learn about the latest progress in microscopic technology, I organized a symposium and invited Ernst Stelzer, who developed the light sheet microscope, and asked him for suggestions on how to improve spatial resolution using light sheet technology. He told me that I might be able to do it with a Bessel beam. From a literature search, I found out that Eric Betzig had developed a novel high-resolution microscope using the Bessel beam technique.
In 2012, Yasushi Okada from [itg-tooltip tooltip-content=”<p><strong>QBiC</strong> is the <a href=&aquot;http://www.qbic.riken.jp/english/&aquot;>RIKEN Quantitative Biology Center</a></p>”]QBiC[/itg-tooltip] invited Eric to an annual meeting of the Japan Society of Cell Biology. At the beginning of his talk on the Bessel beam microscope, he used my EB1-GFP movie (a conventional 2D one taken around 1999) to show the dynamic nature of cell interiors. I was filled at excitement at the prospect of being able to see the EB1-GFP movie in 3D, but in the end he did not present that data during the talk. When I asked him why, he explained that indeed they had tried to collect EB1-GFP motion in 3D, but had failed to do it because they couldn’t prepare nice cell samples. So I shared my HeLa cells expressing EB1-GFP.
About a year later, I got an email telling me that they had succeeded in collecting EB1-GFP 3D movies by developing a new type of microscope. Because the frame rate of the Bessel beam microscope was insufficient, they needed to improve their microscope by creating multiple Bessel beam lines. This was the “lattice light-sheet microscope.”
Finally I was able to obtain 3D EB1-GFP movies, but then I realized that I need to establish a method for analyzing this enormous quantity of multi-dimensional data to extract biologically important information from the data. To do this, I started collaborating with Hideo Yokota’s team in [itg-tooltip tooltip-content=”<p><strong>RAP </strong>is the RIKEN Center for Advanced Photonics</p>”]RAP[/itg-tooltip] and established a computational method for mathematical analysis.
I believe you are planning to build one of these new microscopes yourself. How is the construction going?
So far, I have been able to procure some of the pieces that we will need. How soon we can complete it really depends on budget, but actually it will not cost very much more than a conventional confocal microscope as it will be about 60 million Japanese yen. It is quite exciting, because today there are only three or four of these microscopes in operation, all in the United States. It will be wonderful if we can have one at RIKEN very soon.
How do you plan to use the new technique?
Using this technique, I’m currently analyzing the role of an important tumor suppressor protein whose dysfunction leads to cancer by inducing chromosome instability. The precise molecular mechanism underling the chromosome instability remains to be clarified.
Do you hope that this will lead to therapies for cancer?
Yes, I hope so. Understanding how the microtubules guide the chromosomes and what is disrupted in cancerous cells could allow us to identify targets for drugs that could stop the abnormal cell division. I hope that within the next two or three years that we can identify targets and then begin to move them to the translational phase.
Why do you think the paper was chosen?
Well obviously the fact that it is an important new instrument is important. But actually, many new techniques are developed every year, so I think a key factor is that we were able to actually image these cellular processes and even tissues, and thus show with dramatic images how the microscope could actually be put into use.
Thank you for letting us take up your time today!
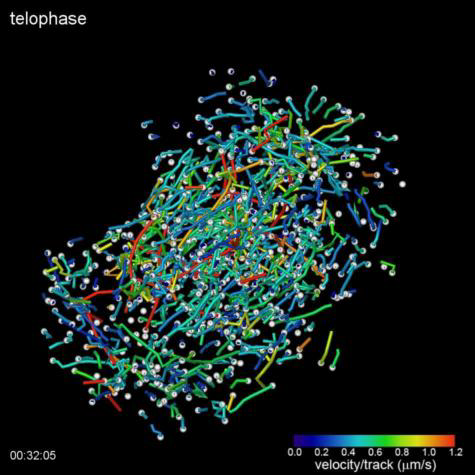
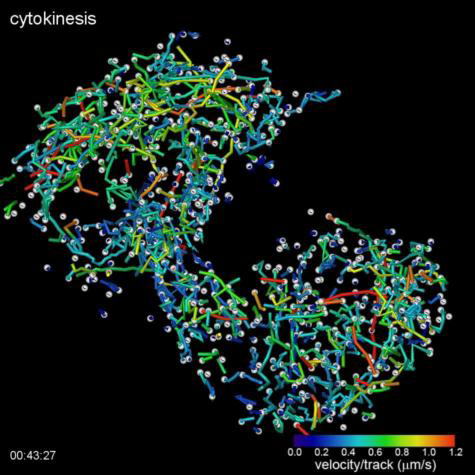
(Top Video) Five different stages during cell division of a HeLa cell as visualized by the lattice light sheet microscope. Measured GFP-EB1 trajectories denote growing microtubule endpoints.
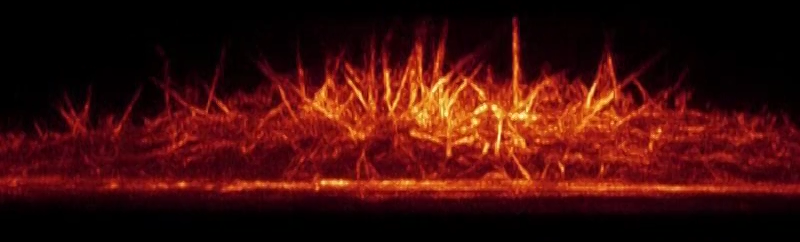
(Bottom Video) Top and side view volume renderings of filopodia in a HeLa cell expressing mEmerald-Lifeact. The high speed of the dithered mode of lattice light-sheet microscopy (left) is compared against the high resolution of the SR-SIM mode (right).
Credits: Betzig Lab, HHMI/Janelia Research Campus; Mimori-Kiyosue Lab, RIKEN Center for Developmental Biology