Fewer components are needed for chromosome assembly during cell division than previously supposed
In a discovery that could lead to the rewriting of biology textbooks, RIKEN researchers have found that tightly wound strands of DNA, known as chromosomes, can form even in the near absence of substructures that were thought to be essential for their production.
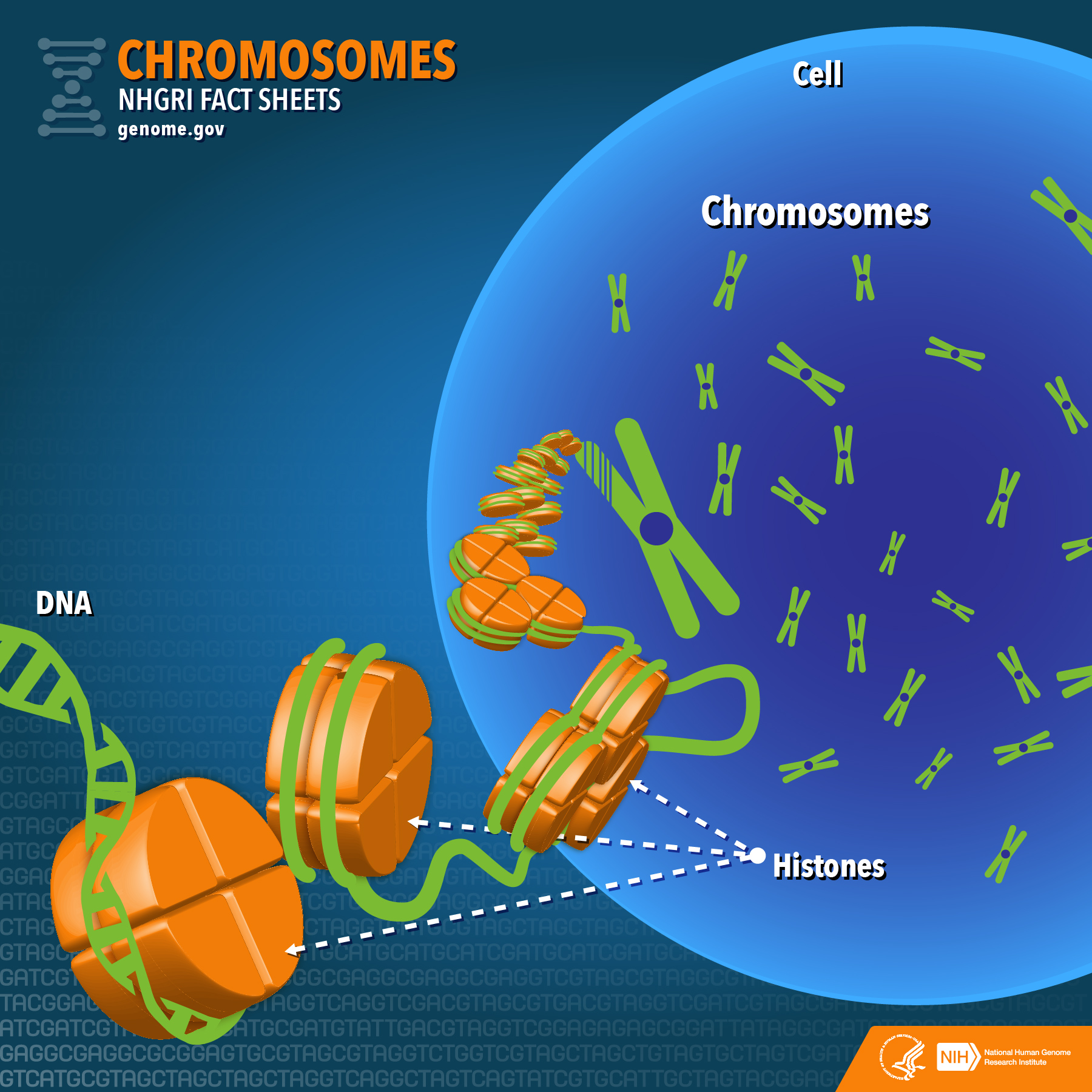
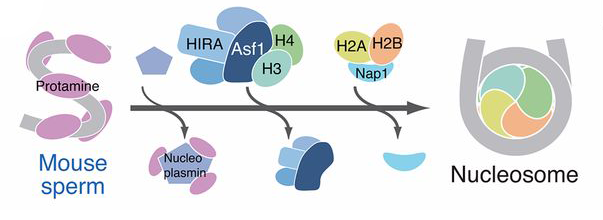
Schematic showing how DNA condenses around histone proteins to form chromosomes
Credit: National Human Genome Research Institute (USA) .
Before a cell divides to form two daughter cells, its DNA is packed into chromosomes. But the exact mechanism for this process is still unknown. “It’s one of the oldest and most fundamental problems in cell biology,” says Tatsuya Hirano, who heads the Chromosome Dynamics Laboratory at RIKEN.
Textbooks on cell biology present a model in which chromosomes assemble in a hierarchy that starts from the assembly of ‘beads-on-a-string’ structures on DNA, known as nucleosomes. Two decades ago, Hirano discovered condensins—large protein complexes responsible for the higher-order folding of nucleosome fibers. It was thus thought that nucleosome assembly underlies higher-order chromosome assembly mediated by condensins.
To test this supposition, Hirano’s team used a ‘cell-free’ experimental system in which chromosomes can be assembled by mixing sperm DNA with an extract prepared from frog eggs that contains all the ingredients to build chromosomes. “We simply mimicked fertilization in a test tube,” explains Hirano. In the current study, the team mixed mouse sperm DNA that lacks nucleosomes with an extract depleted of a protein needed for nucleosome assembly (called Asf1). In this way, just the nucleosome assembly step could be blocked, leaving the other steps intact, including those mediated by condensins.
“We thought that we wouldn’t see anything because many people, including us, believed that nucleosome assembly was a prerequisite for the large-scale organization of mitotic chromosomes,” recalls Hirano.
To the team’s great surprise, the experiment produced chromosomes, although they were somewhat diffuse and fragile. “We were all very excited by that. It may require revising the standard textbook view.”
The team then tested the effect of depleting condensins while interfering with nucleosome assembly. Simultaneous depletion of Asf1 and condensin I had no big effect, whereas depletion of Asf1 and condensin II together led to severe defects in the chromosomes’ architecture. The condensin complexes are already known to act on DNA independently of the nucleosome, at least partially; the new findings hint at complex functional crosstalk between the different components involved in chromosome assembly.
Hirano hopes his lab will eventually unravel the biochemical and biophysical mechanisms of these interactions and how they drive chromosome assembly. “Because I was one of the discoverers of these protein complexes, I feel responsible for fully describing their activity or function,” he comments. ?
[vifblike]
This article originally appeared here as a RIKEN Research Highlight.
In a discovery that could lead to the rewriting of biology textbooks, RIKEN researchers have found that tightly wound strands of DNA, known as chromosomes, can form even in the near absence of substructures that were thought to be essential for their production.

Schematic showing how DNA condenses around histone proteins to form chromosomes
Credit: National Human Genome Research Institute (USA) .
Before a cell divides to form two daughter cells, its DNA is packed into chromosomes. But the exact mechanism for this process is still unknown. “It’s one of the oldest and most fundamental problems in cell biology,” says Tatsuya Hirano, who heads the Chromosome Dynamics Laboratory at RIKEN.
Textbooks on cell biology present a model in which chromosomes assemble in a hierarchy that starts from the assembly of ‘beads-on-a-string’ structures on DNA, known as nucleosomes. Two decades ago, Hirano discovered condensins—large protein complexes responsible for the higher-order folding of nucleosome fibers. It was thus thought that nucleosome assembly underlies higher-order chromosome assembly mediated by condensins.
To test this supposition, Hirano’s team used a ‘cell-free’ experimental system in which chromosomes can be assembled by mixing sperm DNA with an extract prepared from frog eggs that contains all the ingredients to build chromosomes. “We simply mimicked fertilization in a test tube,” explains Hirano. In the current study, the team mixed mouse sperm DNA that lacks nucleosomes with an extract depleted of a protein needed for nucleosome assembly (called Asf1). In this way, just the nucleosome assembly step could be blocked, leaving the other steps intact, including those mediated by condensins.
“We thought that we wouldn’t see anything because many people, including us, believed that nucleosome assembly was a prerequisite for the large-scale organization of mitotic chromosomes,” recalls Hirano.
To the team’s great surprise, the experiment produced chromosomes, although they were somewhat diffuse and fragile. “We were all very excited by that. It may require revising the standard textbook view.”
The team then tested the effect of depleting condensins while interfering with nucleosome assembly. Simultaneous depletion of Asf1 and condensin I had no big effect, whereas depletion of Asf1 and condensin II together led to severe defects in the chromosomes’ architecture. The condensin complexes are already known to act on DNA independently of the nucleosome, at least partially; the new findings hint at complex functional crosstalk between the different components involved in chromosome assembly.
Hirano hopes his lab will eventually unravel the biochemical and biophysical mechanisms of these interactions and how they drive chromosome assembly. “Because I was one of the discoverers of these protein complexes, I feel responsible for fully describing their activity or function,” he comments. ?
[vifblike]
This article originally appeared here as a RIKEN Research Highlight.
Shintomi K, Inoue F, Watanabe H, Ohsumi K, Ohsugi M, Hirano T (2017). Mitotic chromosome assembly despite nucleosome depletion in Xenopus egg extracts. Science. 356:1284–1287. doi: 10.1126/science.aam9702
Shintomi K, Inoue F, Watanabe H, Ohsumi K, Ohsugi M, Hirano T (2017). Mitotic chromosome assembly despite nucleosome depletion in Xenopus egg extracts. Science. 356:1284–1287. doi: 10.1126/science.aam9702
[yuzo_related]